Introduction
In recent years, large language models (LLMs) have displayed extraordinary capabilities across a range of natural language processing tasks, including question answering (QA), text summarization, and conversation modeling. These models, trained on massive corpora of unstructured text, often excel at generating fluent, coherent responses. However, when faced with queries that require complex reasoning, domain-specific understanding, or factual grounding outside their training distribution, LLMs frequently exhibit limitations such as hallucinations, superficial reasoning, and inaccuracies. As the complexity of user queries and domains continues to increase—spanning specialized fields like biomedicine, finance, and scientific literature—there is a growing need for methodologies that can augment LLMs with reliable, context-rich, external knowledge sources.
Knowledge graphs (KGs) offer a structured representation of entities and their relationships, capturing intricate semantic links that go beyond the linear narrative found in unstructured text. By encoding real-world facts and domain-specific information as graph-structured data, KGs provide a natural substrate for advanced reasoning. Despite their potential, integrating KGs into LLM workflows remains challenging. Conventional retrieval-augmented generation (RAG) approaches rely on appending retrieved text documents to the LLM’s input, which can supply facts but not the relational structure inherent in a knowledge graph. Directly bridging the latent space of language models with the structured embeddings of a knowledge graph presents an open research question: How can we inject KG-derived embeddings into an LLM in a way that improves factual grounding, leverages relational information, and preserves the model’s existing linguistic capabilities?
Recent advances in graph representation learning—particularly the success of Graph Neural Networks (GNNs), such as Graph Convolutional Networks (GCNs), Graph Attention Networks (GATs), and GraphSAGE—provide a pathway to encoding and refining node representations that incorporate the topological and semantic structure of KGs. GNNs translate graph connectivity patterns and node attributes into vector embeddings, making it possible to align structured graph knowledge with the linguistic representations in LLMs. However, many existing methods either fine-tune the LLM parameters jointly with graph embeddings or rely on intricate pipelines that compromise the original model’s training distribution.
In this work, we propose a method that integrates GNN-derived embeddings from a knowledge graph into a frozen LLM, specifically a pretrained GPT-2 model. Instead of altering the LLM’s parameters, we treat the GNN and the KG as an adaptable external knowledge engine. For each question-answering scenario, we compute embeddings for both the candidate answers and the relevant node representations from the KG. By selecting and injecting the most appropriate graph-derived embedding directly into GPT-2’s input context, we introduce a latent hint that guides the LLM toward more grounded and contextually consistent predictions.
We evaluate our approach on multiple datasets, including BioGraph—a large, diverse graph sourced from academic literature—and MetaQA, a smaller, domain-specific movie ontology. Our experiments show that GNN-based embeddings integrated into a frozen LLM can significantly improve validation performance on multiple-choice QA tasks. Moreover, the choice of GNN architecture matters: GraphSAGE excels in the larger, sparser BioGraph, while GCN and GAT outperform GraphSAGE on the more structured, domain-specific MetaQA dataset. This variation underscores the importance of tailoring graph representation methods to the nature of the underlying knowledge graph.
Key Novelty
Unlike previous approaches that fine-tune or heavily modify the LLM, our method introduces a simple, direct mechanism for injecting GNN-derived embeddings into a frozen LLM’s latent space. By using a single special token replaced with a GNN-generated embedding, we preserve the integrity of the pretrained language model while enabling it to leverage external graph structure and domain-specific information with minimal overhead, creating a modular and flexible framework for integrating external knowledge.
Methods
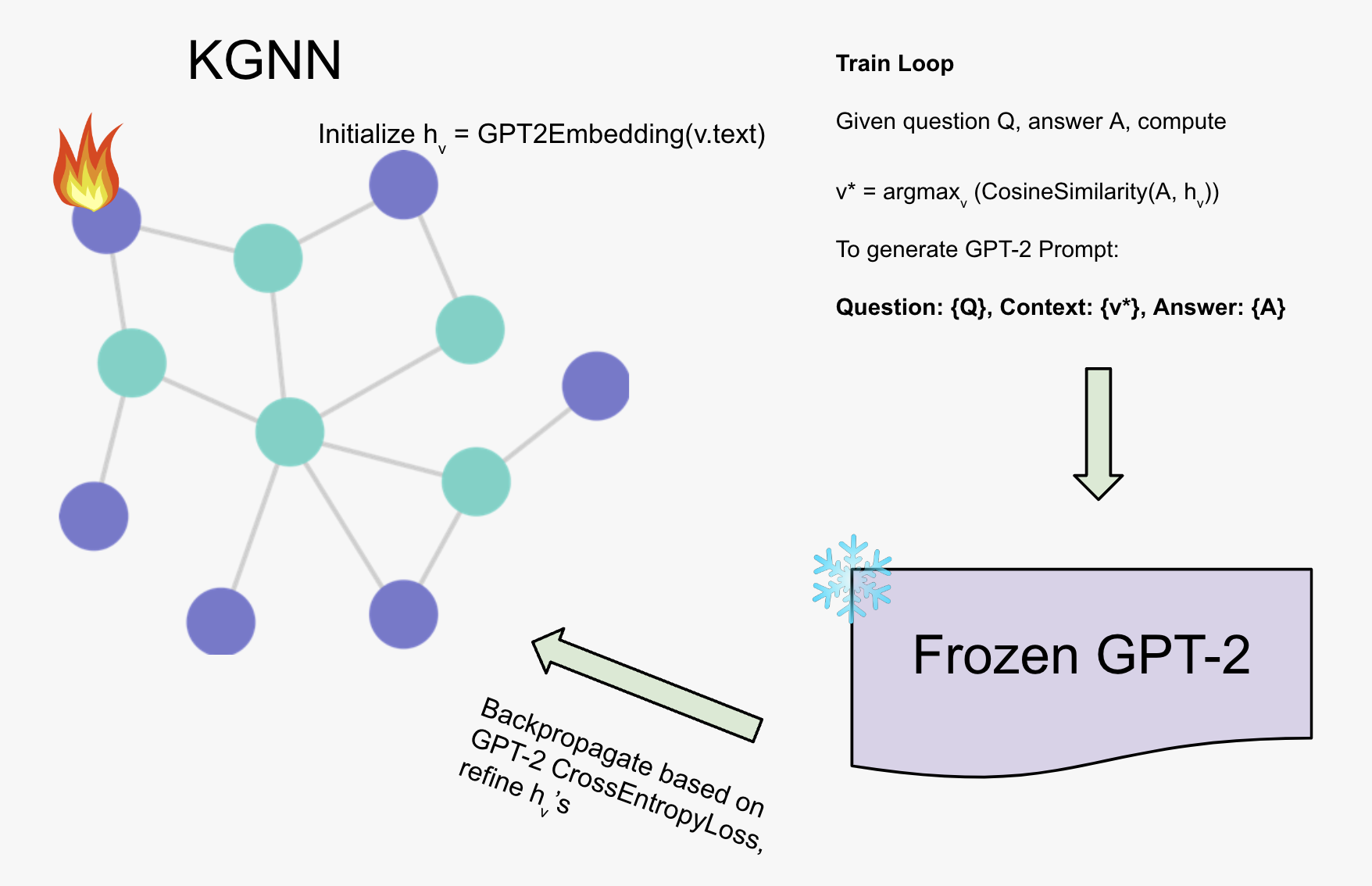
We propose a method to integrate external knowledge from a domain-specific knowledge graph into a large language model (LLM) inference for multiple-choice question answering (QA). This approach employs a pretrained GPT-2 model [1] as a text encoder and a Graph Neural Network (GNN) to refine node embeddings from the knowledge graph. The node features are transformed from text into a token llm, formed by taking the sequence dimension mean of the last layer of GPT-2. By incorporating graph-derived embeddings into GPT-2 prompts, we guide the language model toward knowledge-consistent answers without fine-tuning GPT-2 itself. We train the system as a whole, where the LLM weights are frozen, using its cross entropy loss as a signal for updating the GNN parameters. Figure 1 illustrates the model architecture.
Dataset Characteristics
We conduct our experiments on two datasets: BioGraph, a knowledge graph assembled from academic papers, and MetaQA, a smaller movie-ontology dataset [7]. Table 1 summarizes key structural properties of both datasets.
| Property | Graph Properties (BioGraph) | Graph Properties (MetaQA) |
|---|---|---|
| Number of Nodes | 12319 | 4231 |
| Number of Edges | 15752 | 6712 |
| Average Node Degree | 2.56 | 3.17 |
| Maximum Node Degree | 171 | 98 |
| Minimum Node Degree | 1 | 1 |
| Median Node Degree | 1 | 2 |
The BioGraph dataset is provided as a pre-defined GraphML file, which includes a train and validation split of question-answer pairs referencing nodes in the graph. We load this GraphML file using NetworkX and extract node-level textual descriptors for each entity. For MetaQA, the input format is a list of triple-based facts provided in a plain text file. We parse this file to construct the corresponding graph structure, ensuring consistency with the BioGraph processing pipeline.
Knowledge Graph Embeddings
We start with a domain-specific knowledge graph stored in GraphML format, encoding entities as nodes and their relationships as edges. The graph is loaded using NetworkX [3]. Each node \((i)\) has a textual descriptor \((T_i)\). We tokenize \((T_i)\) and obtain token-level representations from GPT-2’s final-layer hidden states. By averaging this embedding along the sequence dimension, we form the initial node embedding:
$$ h_i = \text{mean}_{t \in T_i} \text{GPT2}(t) $$
This process yields a semantic representation for each node, derived purely from GPT-2’s pretrained language modeling capabilities. GPT-2 parameters remain frozen to preserve the knowledge learned during pretraining.
Graph Neural Network Architectures
After obtaining initial node embeddings \(\{h_i\}\), we consider three GNN architectures to incorporate graph structure: Graph Convolutional Networks (GCN) [2], Graph Attention Networks (GAT) [4], and GraphSAGE [5]. We apply each GNN to the initial embeddings and compare their downstream validation loss.
Graph Convolutional Network
Let \((A)\) be the adjacency matrix of the graph (with self-loops) and \((D)\) the corresponding degree matrix. We initialize \((X^{(0)} = [h_1; h_2; \ldots; h_N])\). A two-layer GCN transforms node features as follows:
$$ X^{(1)} = \sigma(\tilde{D}^{-\frac{1}{2}}\tilde{A}\tilde{D}^{-\frac{1}{2}} X^{(0)}W^{(0)}), \quad X^{(2)} = \tilde{D}^{-\frac{1}{2}}\tilde{A}\tilde{D}^{-\frac{1}{2}} X^{(1)}W^{(1)} $$
Here, \(\tilde{A} = A + I\) and \(\tilde{D}\) is the degree matrix of \(\tilde{A}\). The matrices \((W^{(l)})\) are learnable parameters, and \(\sigma\) is a ReLU activation function. This process produces refined embeddings \((X^{(2)} = [g_1; g_2; \ldots; g_N])\), where each \((g_i)\) encodes the semantic information of node \((i)\) along with its local graph structure.
Graph Attention Network
Graph Attention Networks (GAT) [4] introduces self-attention mechanisms to graph node feature aggregation. Instead of uniformly averaging neighbor information, GAT computes attention coefficients:
$$ \alpha_{ij} = \frac{\exp(\text{LeakyReLU}(a^\top [W^{(0)}h_i \parallel W^{(0)}h_j]))}{\sum_{k \in \mathcal{N}(i)} \exp(\text{LeakyReLU}(a^\top [W^{(0)}h_i \parallel W^{(0)}h_k]))} $$
Here, \(\mathcal{N}(i)\) denotes the neighbors of node \(i\), \(\parallel\) denotes concatenation, \(W^{(0)}\) and \(a\) are learnable parameters. By using multiple attention heads and averaging their outputs, GAT refines node embeddings according to the relevance of each neighbor.
$$ X^{(1)} = \big\Vert_{m=1}^M \sigma\left(\sum_{j \in \mathcal{N}(i)} \alpha_{ij}^{(m)} W^{(0)}h_j\right) $$
Applying another GAT layer similarly yields \(X^{(2)}\), producing attention-enhanced node embeddings.
GraphSAGE
GraphSAGE [5] learns an aggregator function to sample and aggregate neighbor information, enabling inductive node embedding generation for large graphs. A typical GraphSAGE layer updates:
$$ h_i^{(1)} = \sigma\big(W^{(0)} [h_i^{(0)} \parallel \text{AGG}(\{h_j^{(0)}: j \in \mathcal{N}(i)\})]\big) $$
where \(\text{AGG}\) can be a mean, LSTM-based, or pooling aggregator. For our experiments, we use a mean aggregator. After a second layer:
$$ h_i^{(2)} = \sigma\big(W^{(1)} [h_i^{(1)} \parallel \text{AGG}(\{h_j^{(1)}: j \in \mathcal{N}(i)\})]\big), $$
we obtain \((X^{(2)} = [g_1; g_2; \ldots; g_N])\) with GraphSAGE-specific embedding structure.
Integrating Graph Embeddings into the Language Model
Consider a multiple-choice QA scenario with a question \((Q)\) and candidate answers \(\{A_1, A_2, \ldots, A_k\}\). For each answer \((A_j)\), we compute:
$$ a_j = \text{mean}_{t \in A_j} \text{GPT2}(t) $$
We then find the node whose refined embedding \((g_{n_j})\) is most similar to \((a_j)\), using cosine similarity:
$$ n_j = \arg\max_i \frac{a_j^\top g_i}{\|a_j\|\|g_i\|} $$
The selected node embedding \((g_{n_j})\) is inserted into GPT-2’s context. We introduce a special token \(\langle\text{graph}\rangle\) into the model’s prompt:
"Question: \((Q)\)
Context: \(\langle\text{graph}\rangle\)
Answer: \((A_j)\)"
GPT-2 uses its pretrained embeddings for all text except \(\langle\text{graph}\rangle\), which we replace with \((g_{n_j})\). Thus, GPT-2 is conditioned on a graph-derived embedding that provides external contextual knowledge. By keeping GPT-2 frozen, we rely on the GNN to learn representations that are directly useful for improving answer selection.
Training and Objective
We train only the GNN parameters (whether GCN, GAT, or GraphSAGE) by maximizing the likelihood of the correct answer. For each training example \((Q, A_{\text{correct}})\):
$$ \mathcal{L}(\theta) = -\sum_{(Q,A_{\text{correct}})} \log p_\theta(A_{\text{correct}} | Q, g_{n_{\text{correct}}}) $$
Here, \((p_\theta)\) is the probability assigned by GPT-2 when the graph embedding is inserted. GPT-2 remains fixed, and \(\theta\) (the GNN parameters) are updated to yield node embeddings that help GPT-2 choose the correct answer more confidently.
Experimental Setup
All models and training procedures are implemented in PyTorch [8], with GPT-2 integrated using HuggingFace Transformers [9]. GCN, GAT, and GraphSAGE layers are implemented using PyTorch Geometric [10], and NetworkX [3] handles graph loading. By comparing how the validation loss evolves for each GNN-based method on BioGraph and MetaQA, we aim to glean insights into the efficacy of graph-based embeddings for knowledge-consistent QA. We use AdamW [6] for optimization, tuning hyperparameters (e.g., hidden dimensions, learning rates) based on validation performance.
Results
Hyperparameter Validation
Before finalizing our model configurations, we performed a series of validation experiments (all on the BioGraph dataset, due to compute limits) to determine appropriate hyperparameters for each of the GNN architectures (GCN, GAT, and GraphSAGE). We conducted controlled searches over hidden dimension sizes and neighbor sampling rates (for GraphSAGE) on the validation sets of our datasets.
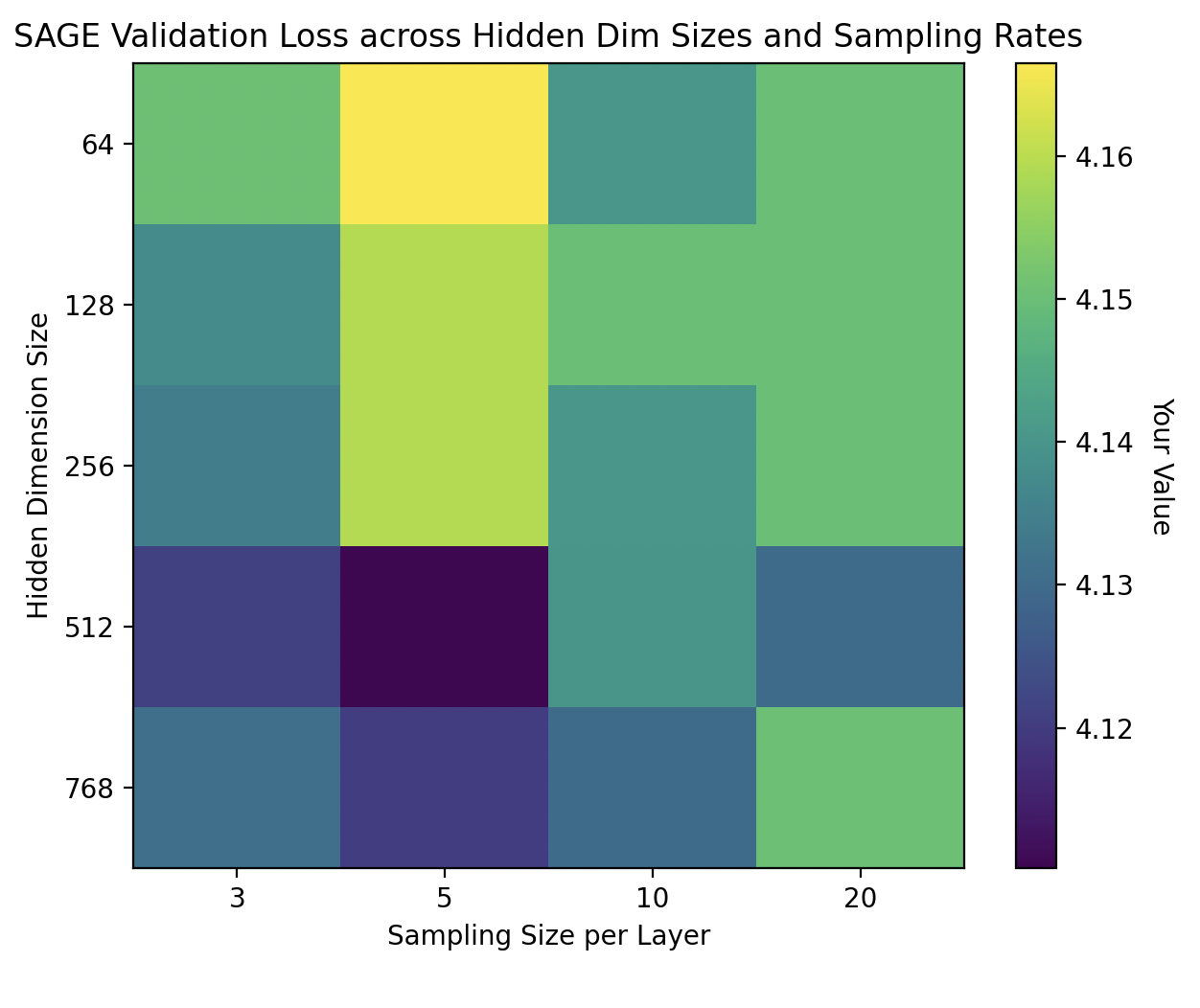
For GraphSAGE, we experimented with hidden dimensions from \(\{64,128,256,512,768\}\) and sampling sizes per layer from \(\{3,5,10,20\}\). As shown in the heatmap below, the optimal configuration for GraphSAGE emerged at a hidden dimension of 512 and a sampling size of 5 neighbors, yielding notably improved validation loss on our validation sets:
For GCN and GAT, we varied the hidden dimensions in the same set \(\{64,128,256,512,768\}\). The validation results revealed that a 512-dimensional hidden layer was also effective for these architectures:
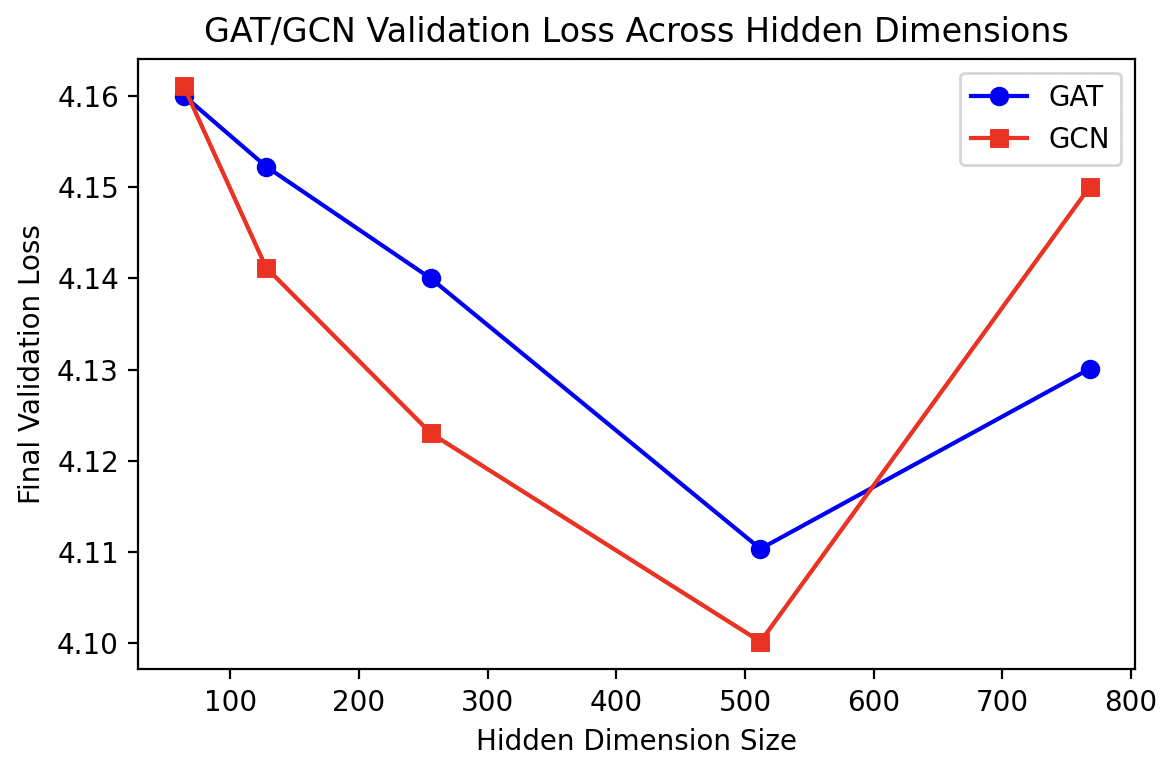
Equipped with these optimized hyperparameters (hidden dimension = 512 for all GNNs and sampling size = 5 neighbors for GraphSAGE), we proceed to evaluate these configurations on both BioGraph and MetaQA datasets.
Performance on BioGraph
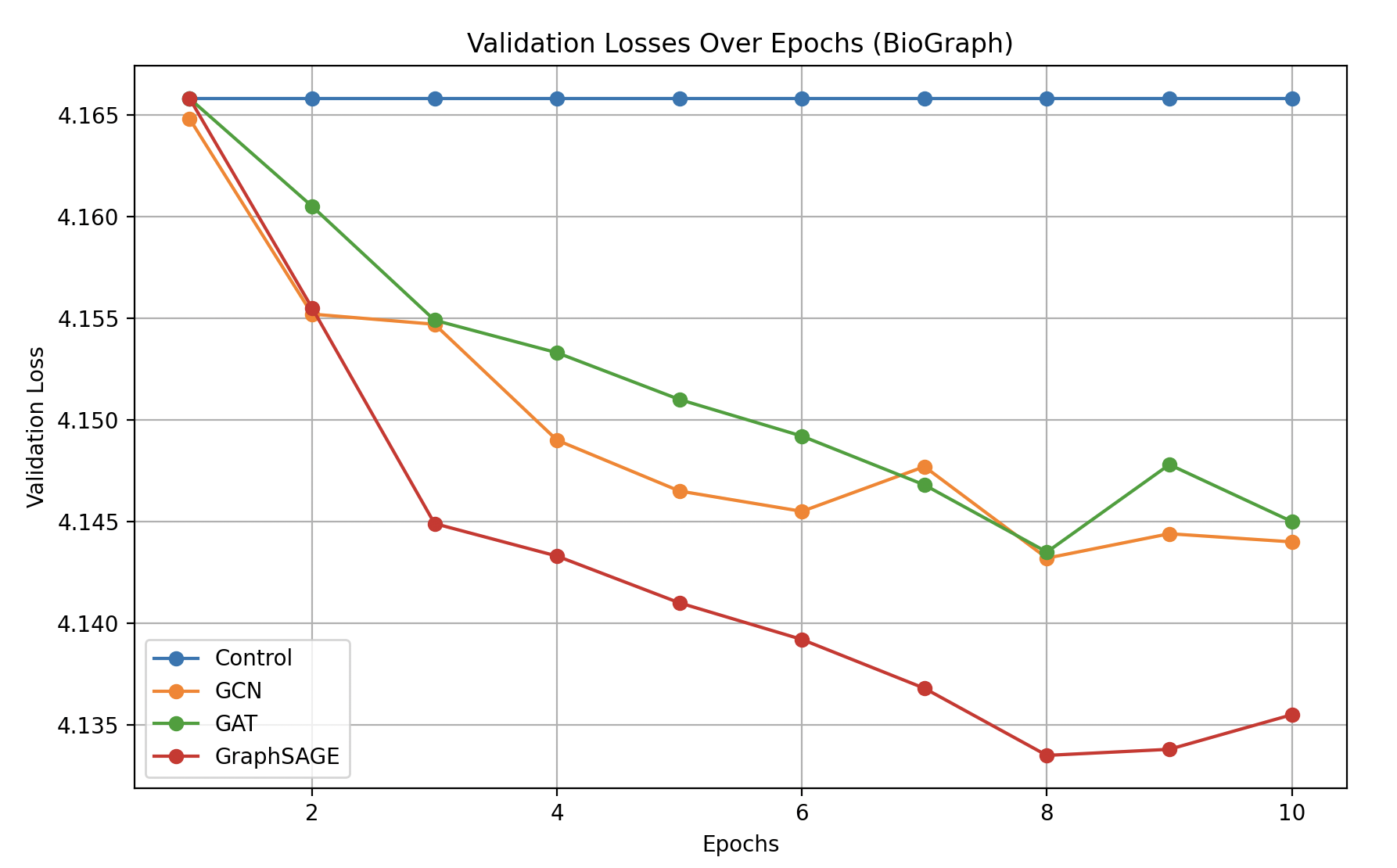
On the BioGraph dataset, integrating graph-derived embeddings into GPT-2 reduces validation loss relative to the control (no GNN) condition. While GCN and GAT yield improvements, GraphSAGE shows the most pronounced reduction in validation loss over epochs. The figure below illustrates how GraphSAGE steadily outperforms both GCN and GAT on this larger, more heterogeneous knowledge graph.
Performance on MetaQA
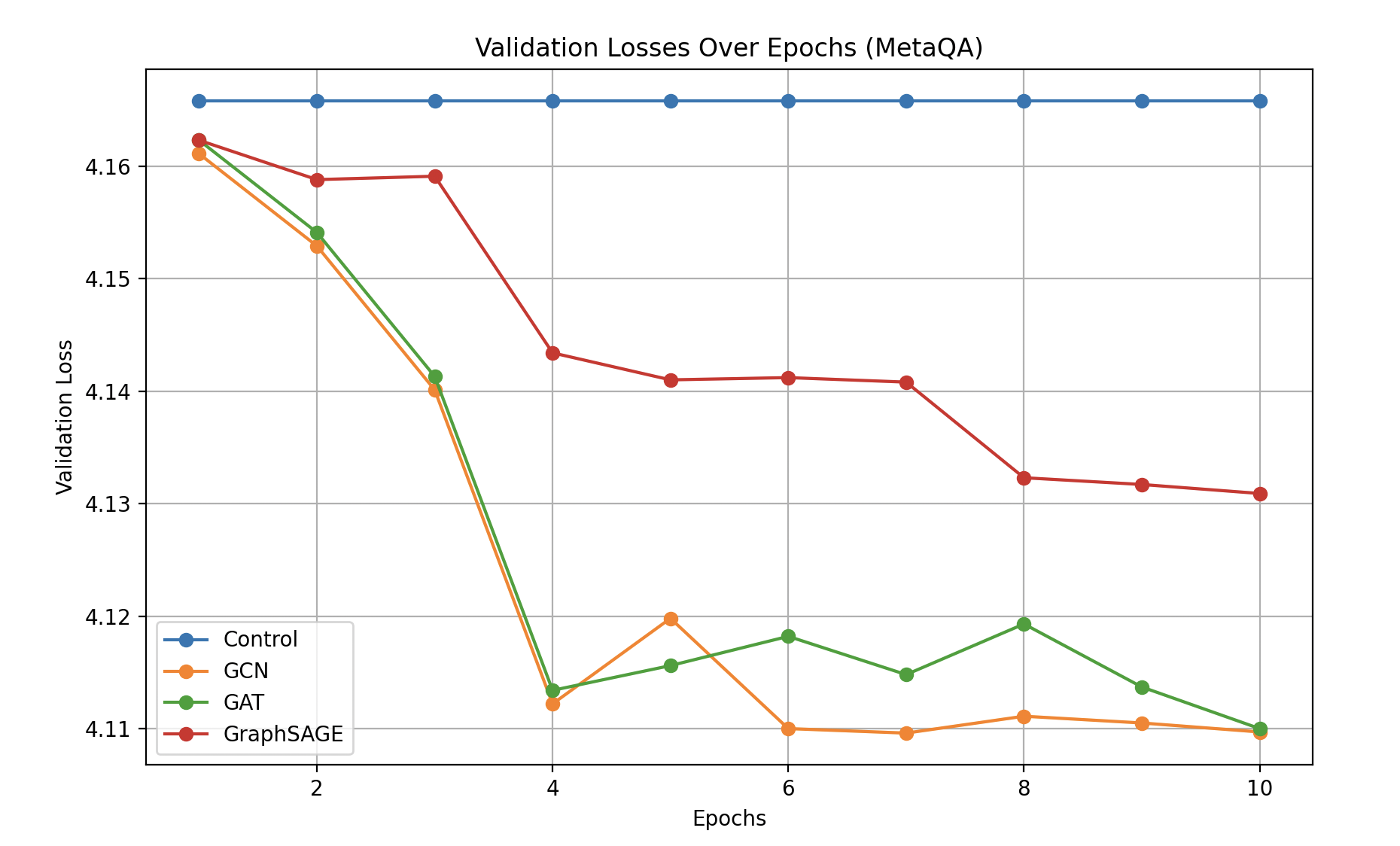
On the smaller MetaQA dataset, all GNN-based methods again outperform the control, confirming that integrating external graph structure is beneficial. However, the relative order of model performance differs from the BioGraph results. Here, GCN and GAT achieve lower validation losses than GraphSAGE, suggesting that the nature of the knowledge graph and its complexity influences which GNN architecture yields the greatest benefit.
The results suggest that while GraphSAGE’s sampling-based approach excels on the larger BioGraph dataset, GCN and GAT might be better suited to MetaQA’s characteristics (e.g., density, connectivity patterns, or domain-specific structure). This highlights that no single GNN architecture universally dominates; performance depends on graph properties and the underlying domain.
Summary of Findings
Our experiments demonstrate that integrating GNN-based embeddings into GPT-2 improves multiple-choice QA performance over a no-graph-control across both BioGraph and MetaQA datasets. However, the best-performing GNN architecture depends on the dataset. On BioGraph, GraphSAGE emerges as the strongest performer, while on MetaQA, GCN and GAT offer superior results.
These observations underscore the importance of selecting and tuning GNN architectures based on dataset characteristics. While 512-dimensional embeddings were generally effective, and GraphSAGE benefited from a sampling size of 5 neighbors, the optimal configuration is task-dependent. Future work may explore more nuanced architectural choices or adaptive approaches that consider graph structure and domain constraints when selecting a GNN.
Discussion
The findings of our study underscore the potential of integrating structured graph-based representations into pretrained language models to improve factual grounding in multiple-choice QA tasks. By leveraging embeddings produced by GNNs—specifically GCN, GAT, and GraphSAGE—and injecting them into GPT-2’s context, we have demonstrated that knowledge graph information can guide a language model towards more informed and contextually relevant answer selection, all while keeping the language model itself frozen.
One key observation emerging from our experiments is the dependency of performance on the characteristics of the underlying knowledge graph and the chosen GNN architecture. Our results show that on BioGraph, a large, heterogeneous graph containing interdisciplinary and sparsely related academic knowledge, GraphSAGE outperformed the other GNN architectures. GraphSAGE was developed with the explicit goal of handling inductive learning settings and efficiently sampling neighborhoods in large graphs. Its success on BioGraph could be attributed to its ability to draw richer, more representative samples from highly varied node neighborhoods. In more complex or less densely connected graphs, adaptive sampling strategies may help retain the most pertinent relational signals. The improved performance of GraphSAGE on BioGraph therefore highlights the importance of carefully considering the nature of the graph and tailoring the GNN choice to its structural properties.
Conversely, on MetaQA—a smaller, more domain-specific dataset focused on movie knowledge—GCN and GAT methods surpassed GraphSAGE. The simpler connectivity patterns and domain-specific ontology of MetaQA may have made standard aggregations (GCN) or attention mechanisms (GAT) more effective. In this environment, sampling strategies may have introduced unnecessary variance or complexity, whereas methods that rely on attentive or uniform aggregation across the entire neighborhood provided more stable and direct representations. This contrast between the two datasets suggests that no single GNN architecture universally outperforms the rest; rather, the “best” GNN often depends on the graph’s density, domain-specificity, and connectivity patterns.
Another important observation is that the improvements in validation loss were more pronounced on the MetaQA dataset. MetaQA’s more cohesive and narrowly scoped domain may offer fewer ambiguous relations, allowing the GNN-augmented language model to more easily form accurate representations. In contrast, BioGraph’s heterogeneous nature introduces complexity that can dilute the immediate impact of graph-derived signals, resulting in more modest, though still valuable, performance gains. This finding underscores the value of domain specificity: more narrowly defined domains, with more coherent and consistent node-to-node relationships, can lead to more substantial improvements when integrating graph embeddings.
While our study demonstrates that integrating GNN embeddings into GPT-2’s prompts can guide the model toward better answer selection, it should be noted that GPT-2 is a relatively small and outdated language model by current standards. This project serves as a proof of concept, illustrating the potential of directly injecting learned node embeddings into a language model’s input stream. Future work should explore more powerful models such as GPT-4 or other state-of-the-art LLMs. With more computational resources and larger models, it may be possible not only to see greater improvements in accuracy and factual grounding but also to rigorously evaluate downstream QA performance, rather than relying solely on validation loss. Moreover, adopting more diverse and sophisticated GNN architectures—or even architectures that dynamically adapt their processing strategy based on the graph’s characteristics—could yield further gains. Temporal or multimodal graphs, where nodes and edges evolve over time or incorporate signals from images, audio, or structured databases, represent another promising avenue for extension.
In sum, our results point toward a research direction where structured knowledge, represented via tailored GNN embeddings, can enhance LLM capabilities without requiring expensive finetuning. As graph data and language models continue to evolve, the synergy between these domains holds the potential to advance our ability to answer complex questions, reduce hallucinations, and ultimately enrich the knowledge integration process in AI-driven information retrieval and decision-making.
References
- Radford, A., et al. (2019). Language Models are Unsupervised Multitask Learners. OpenAI Blog.
- Kipf, T. N., & Welling, M. (2017). Semi-Supervised Classification with Graph Convolutional Networks. ICLR.
- Hagberg, A. A., Swart, P. J., & S Chult, D. A. (2008). Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference (SciPy2008).
- Veličković, P., et al. (2018). Graph Attention Networks. ICLR.
- Hamilton, W., Ying, R., & Leskovec, J. (2017). Inductive representation learning on large graphs. NeurIPS.
- Loshchilov, I., & Hutter, F. (2019). Decoupled Weight Decay Regularization. ICLR.
- Clark, P., et al. (2018). Think you have solved question answering? Try ARC, the AI2 Reasoning Challenge. arXiv preprint arXiv:1803.05457.
- Paszke, A., et al. (2019). PyTorch: An imperative style, high-performance deep learning library. NeurIPS.
- Wolf, T., et al. (2020). Transformers: State-of-the-art natural language processing. EMNLP.
- Fey, M., & Lenssen, J. E. (2019). Fast Graph Representation Learning with PyTorch Geometric. ICLR Workshop.
- Cucala, D. T., et al. (2022). Explainable GNN-based Models over Knowledge Graphs. Department of Computer Science, University of Oxford.
- Mavromatis, C., et al. (2024). GNN-RAG: Leveraging Graph Neural Networks for Knowledge Graph Question Answering. University of Minnesota.
- Li, S. (2021). Understanding Word2vec Embedding in Practice. Towards Data Science.
- Wei, J., et al. (2024). Large Language Model Meets Graph Neural Network in Knowledge Distillation. arXiv preprint arXiv:2402.05894.
- Zhang, Y., et al. (2023). Understanding and Constructing Latent Modality Structures in Multi-modal Representation Learning. Amazon Science.
- Passigan, P. (2023). Continuous Prompt Generation from Linear Combination of Discrete Prompt Embeddings. arXiv preprint arXiv:2312.10323
- Drozd, A., et Al. (2016). Word Embeddings, Analogies, and Machine Learning: Beyond king - man + woman = queen. Proceedings of COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers, 3519-3530.
- Kim, C. (2024). Simple arithmetic operation in latent space can generate a novel three-dimensional graph metamaterials. arXiv preprint arXiv:2404.06671.
- Bhargava, A. (2024). Graph Neural Networks for Knowledge Graphs. University of California, San Diego.
- Li, M.et. al (2024). Simple is effective: Graph-based Knowledge Graph Embeddings for Question Answering. Georgia Institute of Technology.
- Werner, L. et. al (2023). Knowledge Enhanced Graph Neural Networks for Question Answering. Universite Grenoble Alpes, France.
- Arora, S. (2020) A Survey on Graph Neural Networks for Knowledge Graph Completion. Indian Institute of Technology, Delhi, India.